

Posted by Luyao Ma on July 10, 2022
Posted in news
Posted by Luyao Ma on October 31, 2021
Posted in news
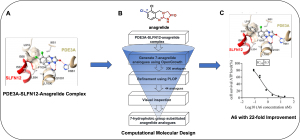 近年来,de Waal、王晓东团队、安然等人相继发现小分子DNMDP、高浓度雌二醇、天然产物nauclefine、PDE3抑制剂anagrelide可以促进PDE3A(phosphodiesterase 3A)和SLFN12(Schlafen family member 12)依赖的细胞凋亡。随后,清华大学王宏伟团队解析了小分子化合物DNMDP,nauclefine,anagrelide结合的PDE3A-SLFN12复合物结构。其中的PDE3A-SLFN12-anagrelide复合物结构显示,结合于PDE3A催化口袋的小分子化合物anagrelide作为分子胶水,“创造”了PDE3A的疏水界面,从而与SLFN12的短螺旋(E552-I558)结合,将两种蛋白质“粘合”在一起,诱导了PDE3A和SLFN12形成稳定的三元复合物。
通过深入分析PDE3A-SLFN12-anagrelide的三元复合物,黄牛团队注意到anagrelide的7位氯原子与PDE3A的T844、F972、L910等残基和SLFN12的I557、I558等残基形成广泛的疏水相互作用(Fig. A)。因此推测,7位氯原子对anagrelide分子胶的作用有重要贡献,对anagrelide凋亡诱导活性也至关重要。基于PDE3A-SLFN12-anagrelide的三元复合物结构和anagrelide 7位氯原子的认知,黄牛团队使用OpenGrowth程序自动对anagrelide 7位的取代基进行采样,期望获得新型的取代基能促进PDE3A和SLFN12之间的相互作用,进而提高小分子化合物的凋亡诱导活性。随后使用MM-GB/SA自由能计算对生成的anagrelide类似物进行亲和力排名。其中排名靠前的以7位取代基为芳环,尤其是苯环或者取代苯环为主。同时,基于对结合模式的理解,团队成员挑选了一批疏水性的取代基(包括苯基)作为优选分子。该计算流程十分高效,一共生成、评价了200个anagrelide 7位取代基的分子,单核10小时内即可完成(Fig. B)。
随后,齐湘兵团队参考计算团队的结果,合成了anagrelide 7 位具有不同疏水性取代的anagrelide类似物,包括芳环或脂肪链。令人鼓舞的是,苯环取代的A4表现出优异的凋亡诱导活性,IC50达到0.56 nM,与anagrelide相比,活性提高了近12倍,甲苯基取代的类似物(A6)取得了最有效的凋亡诱导活性(IC50 = 0.30 nM),比anagrelide提高约22倍(Fig. C)。更重要的是,化合物A6在体内显示出比anagrelide更好的肿瘤生长抑制作用。
该工作由王晓东团队联合王宏伟团队、黄牛团队和齐湘兵团队共同完成。黄牛团队的王元勋博士完成了计算和分子设计的工作。论文发表于Nature Communications杂志上题为
Structure of PDE3A–SLFN12 Complex and Structure-based Design for A Potent Apoptosis Inducer of Tumor Cells
Jie Chen, Nan Liu, Yinpin Huang, Yuanxun Wang, Yuxing Sun, Qingcui Wu, Dianrong Li, Shuanhu Gao, Hong-Wei Wang, Niu Huang, Xiangbing Qi & Xiaodong Wang
近年来,de Waal、王晓东团队、安然等人相继发现小分子DNMDP、高浓度雌二醇、天然产物nauclefine、PDE3抑制剂anagrelide可以促进PDE3A(phosphodiesterase 3A)和SLFN12(Schlafen family member 12)依赖的细胞凋亡。随后,清华大学王宏伟团队解析了小分子化合物DNMDP,nauclefine,anagrelide结合的PDE3A-SLFN12复合物结构。其中的PDE3A-SLFN12-anagrelide复合物结构显示,结合于PDE3A催化口袋的小分子化合物anagrelide作为分子胶水,“创造”了PDE3A的疏水界面,从而与SLFN12的短螺旋(E552-I558)结合,将两种蛋白质“粘合”在一起,诱导了PDE3A和SLFN12形成稳定的三元复合物。
通过深入分析PDE3A-SLFN12-anagrelide的三元复合物,黄牛团队注意到anagrelide的7位氯原子与PDE3A的T844、F972、L910等残基和SLFN12的I557、I558等残基形成广泛的疏水相互作用(Fig. A)。因此推测,7位氯原子对anagrelide分子胶的作用有重要贡献,对anagrelide凋亡诱导活性也至关重要。基于PDE3A-SLFN12-anagrelide的三元复合物结构和anagrelide 7位氯原子的认知,黄牛团队使用OpenGrowth程序自动对anagrelide 7位的取代基进行采样,期望获得新型的取代基能促进PDE3A和SLFN12之间的相互作用,进而提高小分子化合物的凋亡诱导活性。随后使用MM-GB/SA自由能计算对生成的anagrelide类似物进行亲和力排名。其中排名靠前的以7位取代基为芳环,尤其是苯环或者取代苯环为主。同时,基于对结合模式的理解,团队成员挑选了一批疏水性的取代基(包括苯基)作为优选分子。该计算流程十分高效,一共生成、评价了200个anagrelide 7位取代基的分子,单核10小时内即可完成(Fig. B)。
随后,齐湘兵团队参考计算团队的结果,合成了anagrelide 7 位具有不同疏水性取代的anagrelide类似物,包括芳环或脂肪链。令人鼓舞的是,苯环取代的A4表现出优异的凋亡诱导活性,IC50达到0.56 nM,与anagrelide相比,活性提高了近12倍,甲苯基取代的类似物(A6)取得了最有效的凋亡诱导活性(IC50 = 0.30 nM),比anagrelide提高约22倍(Fig. C)。更重要的是,化合物A6在体内显示出比anagrelide更好的肿瘤生长抑制作用。
该工作由王晓东团队联合王宏伟团队、黄牛团队和齐湘兵团队共同完成。黄牛团队的王元勋博士完成了计算和分子设计的工作。论文发表于Nature Communications杂志上题为
Structure of PDE3A–SLFN12 Complex and Structure-based Design for A Potent Apoptosis Inducer of Tumor Cells
Jie Chen, Nan Liu, Yinpin Huang, Yuanxun Wang, Yuxing Sun, Qingcui Wu, Dianrong Li, Shuanhu Gao, Hong-Wei Wang, Niu Huang, Xiangbing Qi & Xiaodong Wang Posted by Luyao Ma on May 11, 2020
Posted in news
2020 年 2 月 25 日,北京生命科学研究所/清华大学生物医学交叉研究院黄牛实验室在《Frontiers in Pharmacology》杂志发表题为“Predicting or Pretending: Artificial Intelligence for Protein-Ligand Interactions Lack of Sufficiently Large and Unbiased Datasets”的文章,发现 AI 用于预测蛋白-配体相互作用时,常用的训练集(PDBbind和DUD-E)对AI模型训练存在严重数据偏向(data bias),导致 AI 模型表现虚高,预测缺乏泛化能力和鲁棒性,误导本领域的方法发展和实际应用,并基于此提出了如何客观评估 AI 模型的观点和建议。
Posted by Luyao Ma on February 12, 2020
Posted in news
Hong Lin, Xiaozhe Zhang, Li Liu, Qiuyu Fu, Chuanlong Zang, Yan Ding, Yang Su, Zhixue Xu, Sining He, Xiaoli Yang, Xiayun Wei, Haibin Mao, Yasong Cui, Yi Wei, Chuanzheng Zhou, Lilin Du, Niu Huang, Ning Zheng, Tao Wang, and Feng Rao